1. Morales, A.E.†, Dong, Y.†, Brown, T., Baid, K., Kontopoulos, D.-G., Gonzalez, V., Huang, Z., Ahmed, A.-W., Bhuinya, A., Hilgers, L., Winkler, S., Hughes, G., Li, X., Lu, P., Yang, Y., Kirilenko, B.M., Devanna, P., Lama, T.M., Nissan, Y., Pippel, M., Dávalos, L.M., Vernes, S.C., Puechmaille, S.J., Rossiter, S.J., Yossi, Y., Prescott, J.B., Kurth, A., Ray, D.A., Lim, B.K., Myers, E., Teeling, E.C., Banerjee, A., Irving, A.T.✉, and Hiller, M.✉ (2023). Reference-quality bat genomes illuminate adaptations to viral tolerance and disease resistance. Research Square. 
Abstract:
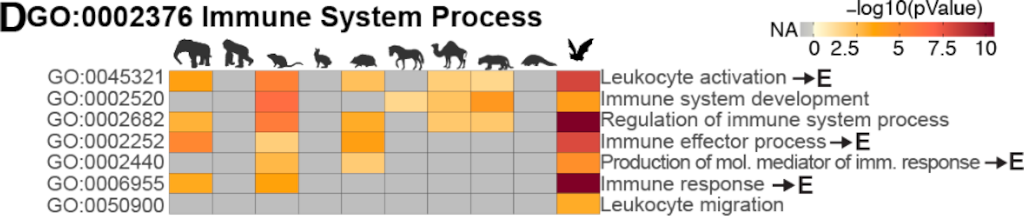
Bats carry viruses that can cause severe disease in other mammals. Asymptomatic infections in bats suggest limited tissue-damaging inflammation and immunopathology. To investigate the genomic basis of disease resistance, the Bat1K project generated reference-quality genomes of ten bat species. A systematic analysis showed that signatures of selection in immune genes are more prevalent in bats compared with other mammals. We found an excess of immune gene adaptations in the ancestral Chiroptera and many descending bat lineages, highlighting viral entry and detection factors, and regulators of antiviral and inflammatory responses. ISG15, an antiviral gene contributing to hyperinflammation during COVID-19, exhibits a deletion of a cysteine, required for homodimer formation, in rhinolophid and hipposiderid bats. Cellular infection experiments showed enhanced intracellular protein conjugation of bat ISG15 and lack of secretion into extracellular space, where human ISG15 stimulates inflammation. Our work highlights molecular mechanisms contributing to viral tolerance and disease resistance in bats.