❝Nothing in biology makes sense except in the light of evolution.❞
Theodosius Dobzhansky, American Biology Teacher, 1973
Climate change is among the most important threats to global biodiversity. To accurately forecast its impacts on the biosphere, it is important to develop a thorough understanding of current and historical responses of diverse biological systems to environmental changes.
Two major themes of my research to date are the following:
How do traits of ectotherms respond to changes in temperature?
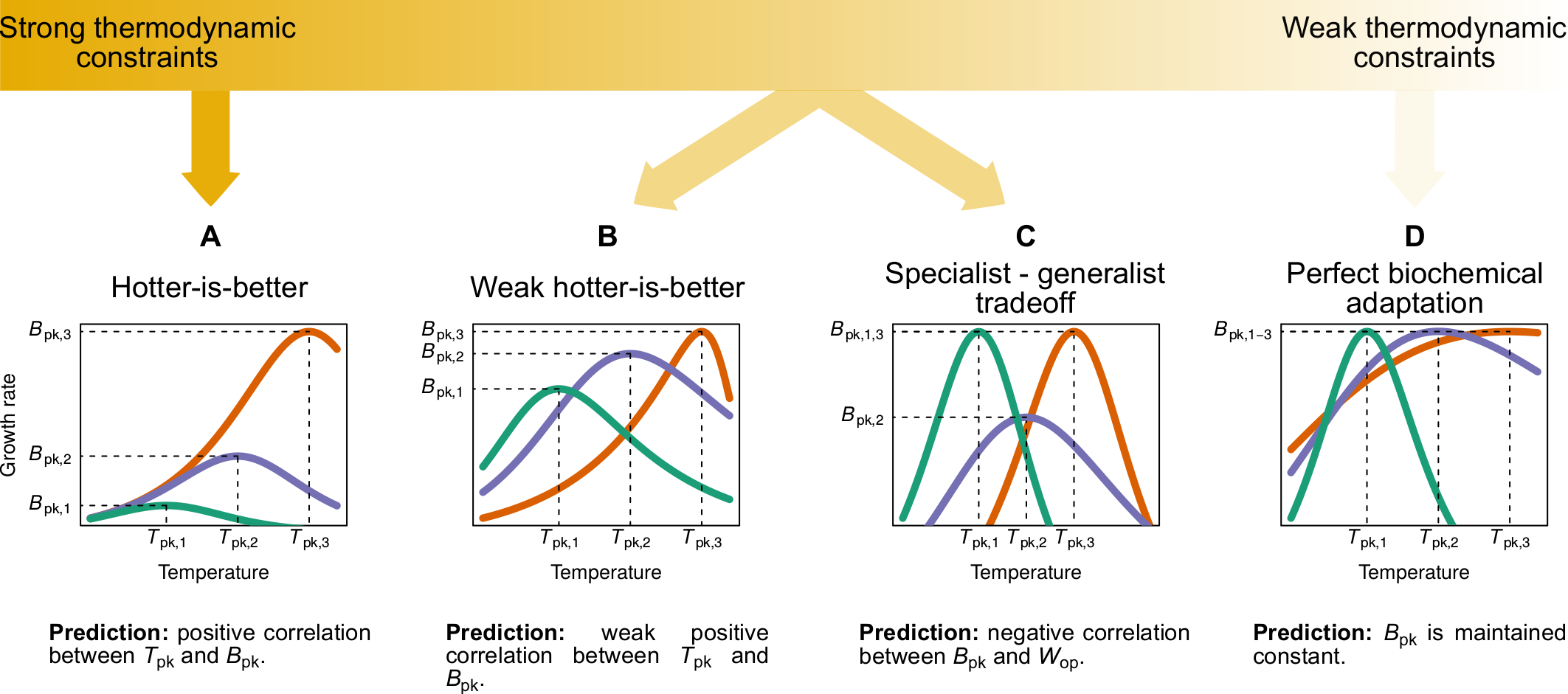
In ectotherms, the performance of physiological, ecological, and life-history traits (e.g., respiration, consumption rate) is typically a unimodal function of temperature. The shape of this relationship exhibits remarkable variation across traits, taxa, and environments. To better understand how such variation emerges, I perform meta-analyses of large empirical datasets, accounting for phylogeny and key environmental factors.
Selected key publications:
What are the physiological, ecological, and genomic underpinnings of torpor in endotherms?

Selected key publications: